Last week we introduced a further function to the move package. It is called the hrBootstrap() which stands for Home Range Bootstrap. It allows to calculate the area of a minimum convex polygon (abbr. MCP) for a set of locations. A polygon is called convex if it includes all line segments connecting any pair of points (see[1]). The minimum convex polygon is the one with the shortest edge length. Given a number of locations of an animal, e.g. due to GPS recordings, the MCP algorithm returns an area that may represent the home range of that animal.
Nowadays the MCP is scarcely if at all used to calculate home ranges. Kernel Density algorithms and Brownian Bridge Movement Models serve more sophisticated results. The results are much closer to the real observations. Nevertheless, the MCP calculation can still be used to compare own results with older studies.
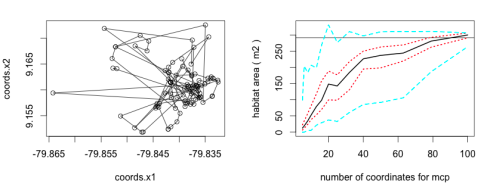
The hrBootstrap() function in our ‘move’ package works for Move and MoveStack objects (individuals or groups of individuals). We implemented here the mcp() and mcp.area() function from the ‘adehabitatHR’ package [2]. We calculate for different number of random locations the mcp. For example for a set of 70 location fixes we calculate the mcp with 5, 6, 8, 10, 13, 16, 20, 25, 32, 40, 50, and 63 locations. Each step is repeated by default 100 times, each time with different locations. The returned data frame represents the calculated mcp area (you can specify the unit with the unout argument) for the according quantiles (0%, 25%, 50%, 75%, 100%) of the values.
By default the quantiles for the different number of locations are plotted as a line plot (see below).
The version will be available, as soon as the r-forge page is back online again.
References
[1] Minimum Conevx Polygon (MCP) http://www.stat.ufl.edu/STA6934/Minimum%20Convex%20Polygon%201%2017%2007.pdf
[2] Home Range Estimation in R: the adehabitatHR Package http://mirrors.fe.up.pt/CRAN/web/packages/adehabitatHR/vignettes/adehabitatHR.pdf

